|
PIs: Fred Vannberg, King Jordan, Augusto Valderrama, José María Satizábal & Adalberto Sánchez
Description:
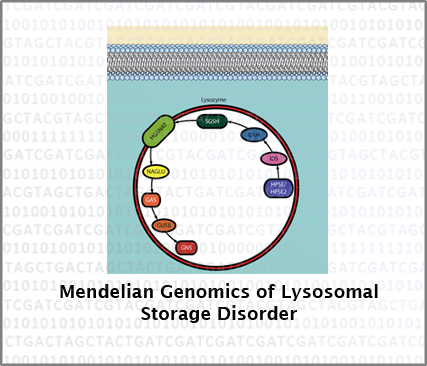
We are working with families from the Valle del Cauca region in SouthWest Colombia that show cases of rare metabolic disorders in order to understand the genomic determinants of Mendelian disease. The emphasis of these studies is on Lysosomal storage disorders, which have a particularly high prevalence in Native American populations from this region.
|